U-Net:Convolutional Networks for Biomedical Image Segmentation
Abstract :
The architecture consists of a contracting path to capture context and a symmetric expanding path that enables precise localization
Introduction :
The convolutional network,their success was limited due to the size of the available training sets and the size of the considered networks.
In many visual tasks,especially in biomedical image processing,the desired output should include localization,i.e.,a class label is supposed to be assigned to each pixel .
Ciresan et al:trained a network in a sliding-window setup to predict the class label of each pixel by providing a local region (patch) around that pixel as input .
- advantages:
- this network can localize
- the training data in terms of patches is much larger than the number of training images
- disadvantages:
- it is quite slow because the network must be run separately for each patch , and there is a lot of redundancy due to overlapping patches.
- there is a trade-off between localization accuracy and the use of context.
- advantages:
More recent approaches proposed a classifier output that takes into account the features from multiple layers.
多尺度融合的深度网络,把某一个像素为中心的不同大小的patch作为多个通道输入到深度网络中学习
In this paper , “fully convolutional network” , the main idea is to supplement a usual contracting network by successive layers , where pooling operators are replaced by upsampling operators .
In the upsampling part we have alse a large number of feature channels , which allow the network to propagate context information to higher resolution layers . The segmentation map only contains the pixels , for which the full context is available in the input image . To predict the pixels in the border region of the image , the missing context is extrapolated by mirroring the input image .
This strategy allows the seamless segmentation of arbitrarily large images by an overlap-tile strategy.
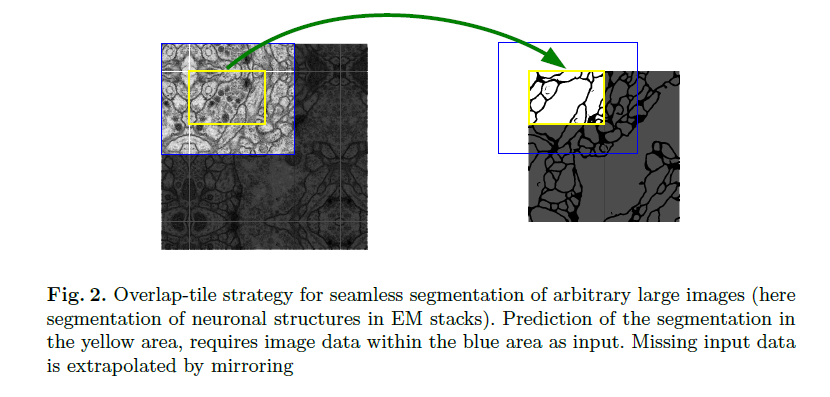
- There is very little training data available , we use excessive data augmentation by applying elastic deformations to the available training images .
- Another challenge is the separation of touching objects of the same class . We propose the use of a weighted loss , where the separating background labels between touching cells obtain a larger weight in the loss function
Network Architecture
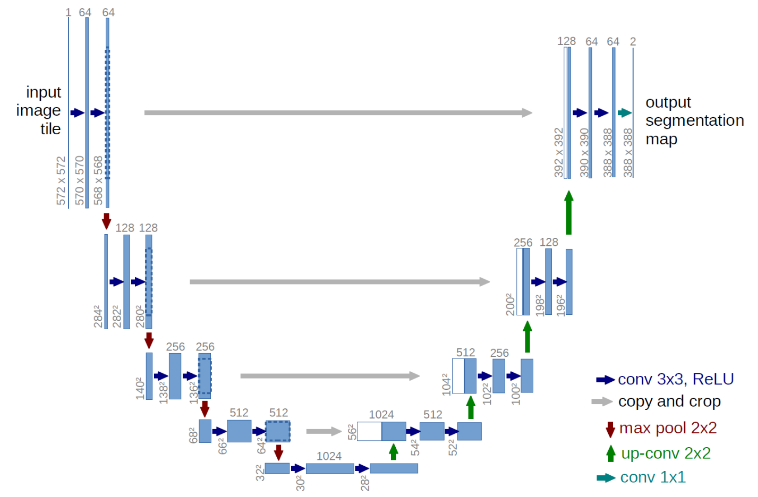
Training
SGD
unpadded
larger input tiles over a large batch size
softmax
The separation border computed using morphological operations
中间项链的部分像素,人为的提高权重,让network可以重点学习这些特征,以便于分割
draw the initial weights from a Gaussian distribution
Data Augmentation
Data Augmentation is essential to teach the network the desired invariance and robustness properties , when only few training samples are available .
- Need shift and rotation invariance as well as robustness to deformations and gray value variations
- Generate smooth deformations using random displacement vectors on a coarse 3 by 3 grid .
Experiments
- 30 images(512*512 pixels)
- ground truth
- membranes
- The u-net (averaged over 7 rotated version of the input data)
Conclusion
The u-net architecture achieves very good performance on very different biomedical segmentation applications . Thanks to data augmentation with elastic deformations , it only needs very few annotated images and has a very reasonable training time .